Over one-third of the world's population is infected with Tuberculosis (TB), caused by Mycobacterium tuberculosis.
Currently 2 million people die each year due to this desease, and new infections occur at a rate of one per second. Due to the
rise of multidrug and extensively resistant strains and its synergism with HIV, tuberculosis has reemerged as a serious public
threat. Hence novel and efficient drugs are urgently needed.

Structure Based Drug Design: The iterative process whereby compounds generated by molecular modeling are synthesized and
crystallized with their target protein. The interactions between the lead compound and the protein can then be analyzed
and the knowledge gained by this analysis will be used to synthesize new lead compounds with higher affinity to their target
protein.
The proteins InhA and KasA of the fatty acid biosynthesis pathway are indespensable for the organism and hence could be a promising
drug target against M. tuberculosis. The structural characterization of target proteins in the presence and absence of bound
inhibitor serves as a basis for the Structure Based Drug Design cycle. Subsequently the structures are used for modeling studies
which will result in potential inhibitors.
After synthesis and biological evaluation the improved compounds are yet again co-crystallized with the target protein to investigate
and understand the interactions between inhibitor and protein. Several rounds of this iterative process can finally educe new drug
candidates.
The enoyl ACP reductase InhA is required for the synthesis of mycolic acids. These long chain fatty acids are an integral and essential component of the mycobacterial cell wall. Blocking the function of this enzyme, which catalyzes the last and rate-limiting step of the fatty acid biosynthesis cycle, is one possibility to prevent the synthesis of mycolic acids in M. tuberculosis.
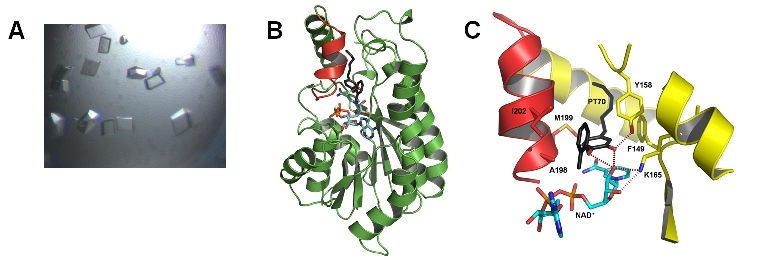
Structure of InhA with bound inhibitor. We co-crystallized InhA in complex with the high affinity inhibitor PT70 and solved the structure of the ternary complex InhA-NAD-PT70. (A) InhA-PT70 crystals. (B) The substrate binding loop (shown in red) was observed in a closed conformation for InhA. The InhA cofactor NAD+ is shown in stick representation. Due to the closure of the binding pocket (zoom shown in (C)) the residence time for the inhibitor (shown in black) is increased. Slow binding inhibitors like PT70 are not released quickly from the target protein and hence provide the basis for a potential application.
Like InhA, KasA is involved in the fatty acid biosynthesis of M. tuberculosis. In the first step of each fatty acid elongation round, the Claisen condensation reaction between malonyl-ACP and acyl-ACP, is catalyzed by this β-ketoacyl synthase. In crystallization studies, we investigated the interactions between KasA and its inhibitor thiolactomycin.
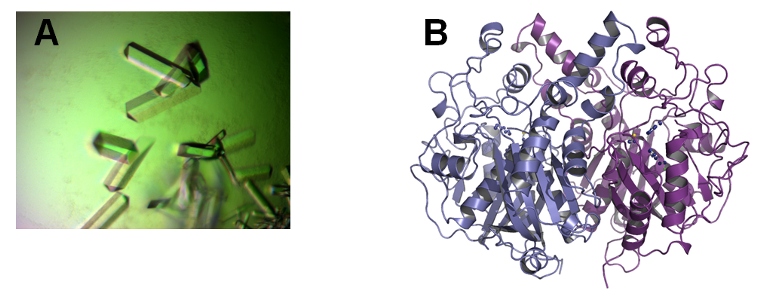
Crystal structure of KasA. (A) KasA crystals. (B) Structural model of the KasA dimer.
Thiolactomycin is a naturally occurring product and shows inhibitory activity towards KasA. Structures of KasA in the presence and absence of thiolactomycin provided first insights regarding the flexibility of the active site and showed which residues are critical for inhibitor binding.
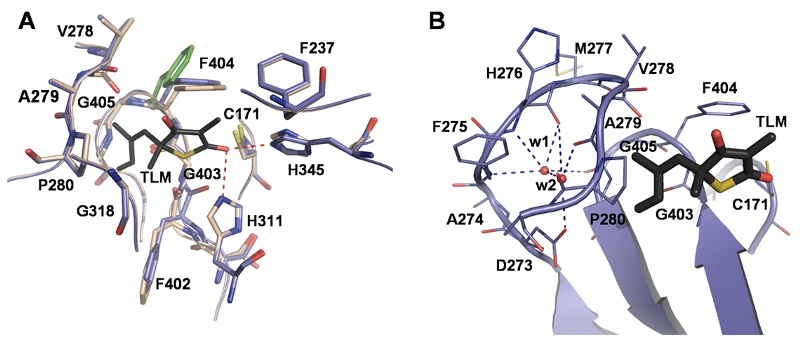
Comparison of crystal structures with and without the inhibitor thiolactomycin. The hydrogen bonds to the active site histidines His345 and His311 contribute to the stabilization of the protein-inhibitor complex. These structures serve as a basis for molecular modeling studies with the aim to obtain inhibitors with improved affinities and bio-availabilities. Our crystal structure of the homo-dimeric KasA has been featured as the structure of the month September 2009 (www.rigaku.com/protein/sotm-011.html).
In addition, we investigate different target proteins involved in proteolysis, fatty acid biosynthesis, vitamin K biosynthesis and cholesterol degradation pathways of various pathogens. Examples for pathogenic organisms that are of interest for us are Trypanosoma brucei, Mycobacterium tuberculosis, Francisella tularensis, Yersinia pestis, Staphylococcus aureus, Burkholderia pseudomallei and the SARS coronavirus. Structures of their proteins with newly developed, promising inhibitors will provide clues for further drug development against infectious diseases.